The World is Her Oyster: Micromollusks are Kind of a Big Deal for Young Scientist
Our Collections and Research Center (CRC) is fertile ground for research. Our curatorial staff and visiting scholars from around the world frequently publish work based on our specimens. Not to brag, but recently a scientist at another small museum—Western Science Center in Hemet, CA—published his discovery of a species of mastodon, based in part on fossil specimens from our Earth Science Collections. Just last week, scientists from a variety of institutions (including two visitors from the Smithsonian Institute’s National Museum of Natural History) studied artifacts in our Anthropology Collections and specimens in our Department of Vertebrate Zoology. Yet the coauthor on one of our recent peer-reviewed publications coming from the Department of Invertebrate Zoology is a little different: she’s a scientist who just happens to be 18 years old.
When Bianca Campagnari joined our work/study/intern program Quasars to Sea Stars, she didn’t know exactly where her path would lead, but she was already interested in molecular biology, and wanted to get involved in research at the Museum. “I really wanted to work here [in the CRC],” she recently recalled. “I thought it was really cool to see all the collections. There’s just so much stuff that we have here, and a lot to learn.”

One example of the CRC’s thousands of wonders: a box of delicate objects that look like shells (or mouthwatering meringues) but are actually egg cases made by the pelagic octopus known as the paper nautilus.
All of our quasars (as the teens in the program are known) intern and work around the Museum and Sea Center. They contribute to the life of the institution in one way or another as they interact with guests, help maintain exhibits, develop educational content, assist in the curation of our collections, or otherwise follow their various passions and talents. Campagnari started working in the Department of Invertebrate Zoology (IZ) because she heard there were many opportunities here, and Curator of Malacology Daniel L. Geiger, Ph.D., was taking on new students to mentor. At that time—about three years ago—Campagnari didn’t have any special fondness for invertebrates. “But now I do,” she admits. It’s one of those things that . . . the more you learn about it, the more you want to learn.”

Campagnari in the IZ lab with a jar of isopods from an Oregon trawl sample (photo by Danial Pirooz). Right, a very graphic sample of diversity in a single drawer from IZ’s Shell Collections.
This insatiable appetite for learning seems common to all our quasars, and in Campagnari, it’s apparent to anyone who hears her discuss her research or gets wind of the extraordinary dedication she’s shown in pursuing it. Campagnari earned her status as primary author on her recent publication* by committing hundreds of hours to tasks that ranged from the tedious to the sophisticated. All that work shines through in this peer-reviewed publication that evaluates four statistical methods for estimating species richness.
Species richness is a count of the number of species present in an area. If you have a sense of species richness, and also a sense of the relative abundance of each species, you have a measure of biodiversity. Biodiversity is important for many reasons, not least of which is the ability of more biodiverse ecosystems to bounce back from crises that might wipe out a more homogenous population. Knowing the species richness and biodiversity of different areas helps governments, NGOs, and scientists decide which areas are most important to study and most efficient to protect. It stands to reason that a coral reef is probably more biodiverse than a sterile kitchen counter, but what about when you’re comparing one coral reef to another? Researchers need practical ways to quantify and compare biodiversity based on sampling at different sites. So Campagnari’s paper employing a group of statistical methods for estimating species richness—the first such analysis applied to micromollusks (extremely small gastropods and bivalves)—has the potential to make a real contribution to conservation efforts.

You can see some species diversity in this photo of a coral reef, but you wouldn’t see any micromollusks in an image taken at this scale. To detect micromollusks, scientists must sample their tiny shells from the sand. Photo by Jim Maragos / U.S. Fish & Wildlife Service
The analysis took as a case study a set of samples from the seafloor in areas off the coast of Maui. The location is interesting because Hawaii’s biodiversity has curious circumstances. Since Hawaii is isolated in the middle of the Pacific, species richness is lower there than in the area around islands in Southeast Asia (where the biodiversity of shelled marine mollusks is highest). Species adapted to coastal conditions that evolved in other parts of the world can’t easily cross vast expanses of deep open water to start a new population in Hawaii. However, because of that isolation, Hawaii is home to many endemics: species that live there and nowhere else, as they have evolved in response to a secluded environment to be genetically distinct from their forebears and relatives elsewhere. This unique natural heritage is worth understanding and protecting.
The Maui samples were donated to the Museum by Hawaiian marine biologist and mollusk expert Mike Severns, long before Campagnari came on the scene. Severns collected the samples by dredging, picking up a big scoop of sediment that included sand and organic matter on the seafloor, but also plenty of micromollusk shells that came along for the ride. The group of shells constituted a thanatocoenosis or “death assemblage.” (Dibs on the band name.) A death assemblage refers to remains collected at a certain site, whether by gravity and ocean currents (as in this case), a catastrophic event, or a predator that creates a refuse heap by bringing meals back to its den.

Would sorting through this bag of sand and tiny shell fragments give you pleasure? You might have a future in micromalacology (the study of tiny mollusks). Photo by Bianca Campagnari
It’s a lot easier to find micromollusks in a death assemblage than on the hoof, as it were. Given their size, they are difficult to find and impossible to identify without access to a microscope. Many are small enough that researchers benefit from viewing them on a scanning electron microscope (SEM) to clearly perceive their distinguishing characteristics. Fortunately, we have one of those, and operating it is one of Dr. Geiger’s specialties. He’s used the SEM to make extensive contributions to the literature on micromollusks (as well as microorchids, another area of expertise).

Dr. Geiger and the SEM. Photo by Chuck Place
When Campagnari embarked on this project, she was the unwitting subject of another scientific inquiry run by Geiger: “When Bianca approached me to do her final project,” he recalls, “I thought, hey, a little human psychology experiment would be fun to do. So I told her, ‘I’ve got this idea. I’ve got this data set with some shells…and I think you could analyze it. There’s this technique called rarefaction. I’ve never done it. And you can use this programming language called R. I’ve never used it. You can figure it out.”
How would Campagnari react to being asked to do not only something she’d never done, but something her mentor had never done either? “As a good scientist, I had a null hypothesis and an alternate hypothesis,” Geiger says. He went into the experiment preferring the null hypothesis, in which Campagnari would have declined the challenge and asked to do something else. His alternate hypothesis, “Much less desirable, because R is a really tricky language—I know that because my wife† uses it—is that maybe she comes back saying ‘You know, I’ve tried to work with it, but I get error 52. What does that mean?’ And that would mean I actually have to understand this, causing me a lot of work and grief, or I have to call in a favor from my wife.”
Geiger gave Campagnari a spreadsheet containing data from some samples he had sorted himself, painstakingly identifying the species present in a bag of undifferentiated sand, shells, and fragments. It was up to Campagnari to research the relevant code, figure out how to run it, prepare the data to be plugged into it, handle any mysterious errors that might arise, and interpret the results. Two weeks later—during their first check-in since beginning the project—“Bianca came back and said, ‘Hey, I made this rarefaction graph in R. What do you think?’ It shocked me,” says Geiger. “An unexpected result! Very exciting from a scientific perspective. She did it all, and in record time.”
In addition to drawing on Campagnari’s early experience with programming, the project allowed her to apply concepts she learned from studying statistics at Santa Barbara City College during her sophomore year of high school. Photo by Claudio Campagnari
Campagnari laughs when she hears her mentor describe how he tested her resolve. “I didn’t think of it as an experiment, I just thought: this is the project, I’ll do it,” she says. “It was a really good challenge.” Although she’d never heard of R at the time Geiger introduced the project, she wasn’t intimidated by the idea of programming, having started some online courses in Python (another programming language) and attended lectures on programming taught by her father, a professor and chair of UCSB’s Department of Physics. Campagnari used Python in her tenth-grade science project.
When Geiger got over his surprise at his experiment’s unexpected result, he examined the graph to see if its projections made sense, and they did. Campagnari took the project a step further by researching other approaches to estimating species richness, and applying those to the same data. They also produced results that made sense, and—best of all—the different methods agreed with one another. “Given how tightly they agreed, it was absolutely stunning. This was a best case scenario,” says Geiger.
So what kind of information do these methods deal in? I won’t—OK, I can’t—describe the statistics behind the estimates in detail, but roughly, they produce curves that show the relationship between the number of individuals in a sample of a certain size and the number of species present in that sample, in a way that is predictive of how many species might be found in a larger sample, and by extension, the area being sampled. The curve rises sharply, but then levels off, showing an effect that makes a lot of sense when Campagnari describes it: “If you were to go to an island, your first day on the island, you’d find a bunch of new species, maybe 100 new species in the first day, but after you spent a long time on the island, you would find new species much less frequently. Maybe 10 years on the island, you would find a new species maybe once a year. So that explains the slope of the graph.” Basically, while you’re probably always going to be finding new species in a given area if you keep sampling more, there’s still a point of diminishing returns. That’s where the curve starts to level off, because a larger sample doesn’t find you many more species.
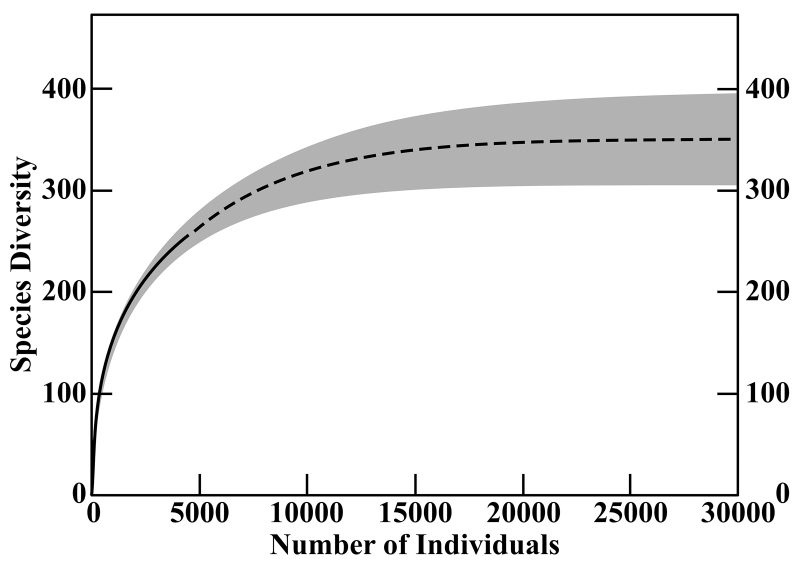
This curve synthesizes the curves produced by three different statistical methods for estimating species richness. The solid line represents the data from 4,233 identified shells. The dashed line represents the predicted richness result of further sampling. The methods agree with 95% confidence that the results would lie within the gray area.
This is especially valuable because the curve is predictive (after all, that’s what makes a function useful and not just pretty to look at): in the specific case of Campagnari’s samples, it shows that based on 4,233 shells identified to 250 species, it would take a sample with about three times as many shells to reach that point of diminishing returns, because there are probably about 350 species to be found in the area. The model’s prediction is rooted in its evaluation of the presence of rare species in the sample (rare being a relative term, in this case meaning rare within the sample). If you’re finding a lot of rare species, the site is probably a biodiversity hotspot. If—to imagine an unlikely opposite extreme—you sample 4,233 shells, identifiable in equal quantities to only two species, it’s definitely not a hotspot, and you should have run your predictive model (or just used your head) a lot sooner! Use cases for these methods lie in more subtle scenarios, as in the question of comparing different coral reefs mentioned above.
With these different methods in agreement, it was a case study worth publishing, both as a trial of these different methods and as an example of how they could be applied to micromollusk samples. The case study came with a bonus: the description of a new species.

The anatomy of newly-described Murdochella tricingulata illuminated by Campagnari’s SEM imagery. Scale bars in images of entire shells are 1 mm, scale bars in detail images are 100 μm.
If you’re wondering why we buried the lede here—What? A newly-described species?—it’s because describing new species is the bread and butter of invertebrate zoology, especially where smaller organisms are concerned. Geiger alone has named over 100 species. That tally includes some orchids, but is mostly made up of tiny invertebrates.
“In general, larger organisms are better known than the small ones,” Geiger explains. “We know about whales and dolphins, but we do not know about mites. That’s also true for mollusks. The larger ones are well known. They have been collected and described, no major surprises there.” This is why Geiger has made a career of using scanning electron microscopy to investigate tiny lifeforms. “In the micro world, once you go below five millimeters…there are lots of discoveries to be made. One of the very basic questions [about these tiny organisms] is ‘How many are out there?’ ” Estimates of species richness can help answer this question.
The process of describing and identifying our planet’s tiny organisms is a time-consuming task with no end in sight. (In our last blog post, we contemplated the vast invertebrate unknown from the perspective of beetle taxonomy.) Unfortunately, we’re altering ecosystems and causing extinctions faster than we can understand the consequences. This means there’s a need to estimate what we don’t know, so we stand a chance of protecting it. We can’t assume that the small size of an organism means it’s ecologically insignificant. Take plankton, for example, the microscopic marine drifters without which our planet’s ecosystems would be unrecognizable. To paraphrase Dr. Seuss, a mollusk’s a mollusk, no matter how small.
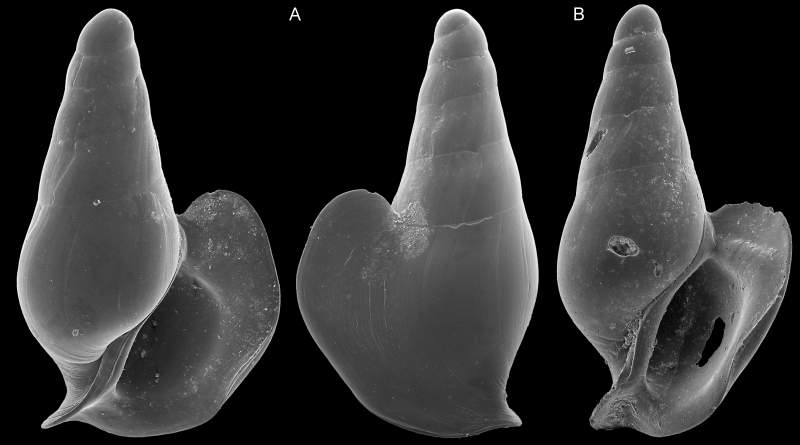
SEM imagery of two specimens used to describe Severnsia strombeulima, named by Geiger in honor of Mike Severns, collector of the samples used in Campagnari’s research and author of extensive literature on mollusks of Hawaii. These shells are less than a millimeter wide. Images by Daniel Geiger
Since the task of discovering and describing these species is still in its early stages, we know very little about their ecological roles. Geiger is the author of the two-volume Monograph of the Little Slit Shells, which—despite its diminutive subjects—is heavy enough to be used as a bludgeon against unwanted intruders. In researching these volumes summarizing the scientific knowledge on six families of micromollusks, he was flabbergasted to find that there was only a single account documenting that a fish had once eaten one individual out of 250 species in this group. “There is one mention in a fish gut analysis that found an unidentified [little slit shell] in a fish gut, and that’s it for the literature on predation.”
Unfortunately, we can’t assume that this means nobody eats these and their importance to the ecosystem is negligible. The biggest problem—aside from how delicate these specimens are, and how unlikely it is that they’ll survive a variety of practical sampling procedures—is that “someone needs to care sufficiently to identify it, and that means a multidisciplinary approach. Usually people are fairly [species]-specific. If it’s a small shell [and you’re focused on a certain fish], whatever, who cares, move on. So this is the state of our knowledge.”
The lack of literature on micromollusks makes Campagnari’s research particularly valuable. It fills a gap; there were lists of micromollusk species known to be present in certain areas, but no studies addressing the problem of estimating species richness for micromollusks. “It’s just hard to work on them, and therefore nobody does the work,” Geiger explains. He first realized these difficulties in the mid-2000s, when he thought he would find a handbook or book chapter on working with micromollusks. When he discovered there was no such guide in existence, he wrote one, in collaboration with colleagues from Australasia, Japan, and Europe.‡
Campagnari made use of that technical knowledge in her work. In sorting one of these Hawaiian samples to produce more data for her analysis, she encountered a problem that researchers studying larger mollusks don’t experience: the difficulty of distinguishing her subjects—tiny shells—from the grains of sand that made up the bulk of the samples. Researchers often use meshes to sort their subjects from the surrounding sediment, but when your subjects are the same size as the particles that surround them, you’re out of luck. Campagnari studied each tiny candidate under a light microscope to see if it was truly a shell. In doing so, she couldn’t always rely on ordinary tweezers to manipulate the tiny, fragile shells. “I had a tray and a light microscope, and different tweezers for different strengths of shells,” Campagnari explains. “A normal pair of tweezers, and then one made for entomologists, and then a paintbrush. If a shell is really thin, you pick it up with the bristles of a paintbrush.” It took 60 hours to sort a single sample, and that was only to winnow the shells from the grains of sand, not to identify them at any taxonomic level.
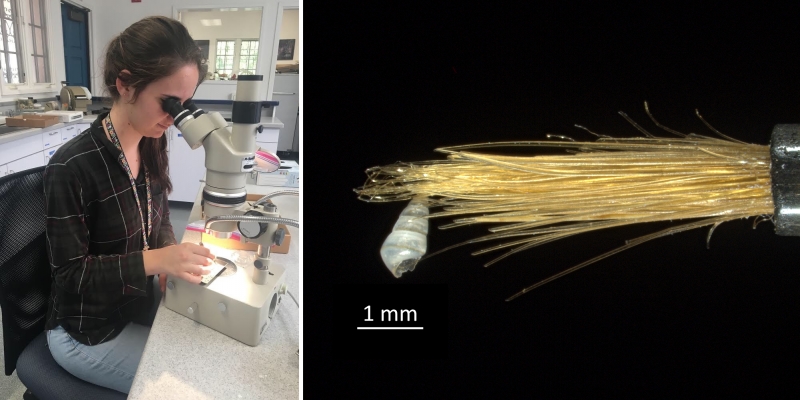
Campagnari sorting shells with the aid of a light microscope. Micromollusks like the tiny specimen at right are extremely fragile. Scientists who work with these delicate specimens employ fine brushes to safely manipulate them. Photos by Daniel Geiger
I asked Campagnari if—given the power to travel back in time and give her past self any advice—she would have made the suggestion not to take the path that would lead to 60 hours of sorting shells from sand as an early step. She said the opposite: “I would tell myself, you actually ended up doing it. Because a lot of times, it was pretty difficult.” She managed her time “by working in really long bursts, just spending all day working” in a highly focused state. She didn’t listen to music. “It’s really easy to focus here, because it’s just a very monotonous task and there’s no noise at all. I get really focused whenever I work here. It’s actually kind of relaxing.” After a long day, she’d take a three-day break from the job, giving her eyes, concentration, and fine motor skills a rest. (In this context, the ordinary life of a busy high school student constitutes a rest.)
One of the many rewards Campagnari received for her labors was the privilege of using the scanning electron microscope to get a substantially closer look at the shells the light microscope could barely distinguish from sand grains. She used the SEM to capture the images used in the publication. Campagnari explains how the SEM is different from an ordinary microscope: “There’s a beam of electrons that goes down and knocks electrons out of the atoms in the sample. Those electrons go to detectors. The amount of electrons reflected depends on the angle the specimen makes with the electron beam, so that steeper surfaces reflect more electrons and translate to brighter zones than flat surfaces.” Together, the detectors can piece together an image revealing the topography of the specimen. This is why SEM imagery is in black and white: it doesn’t represent visible light reflecting in different colors, but is more like a topographical map of a tiny object. The lack of color isn’t important compared to the insight offered by imaging at this astounding level of detail. Without this kind of resolution, it would be impossible to see the distinguishing features that allow experts like Geiger to recognize different species of micromollusks.
Campagnari learned to identify various species over the course of her research. As a predictable science journalist, I asked if she had any favorites, and she expressed a preference for the Epitonidae family, also known as wenteltraps. “Wenteltrap means spiral staircase in some language, so they’re spirally and really round and have nice ridges, and they kind of look like frosting. I like them.” Of course, she’s also partial the new species she and Geiger described in their paper, Murdochella tricingulata. Campagnari had the privilege of naming it, which she used wisely, choosing tricingulata (meaning three-banded) to refer to a visual identifier and complement the previously known species in the genus, Murdochella bicingulata (two-banded).

A wee epitoniid (also known as a wenteltrap), one of Campagnari’s favorite kinds of shells, in the IZ collections. Photo by Bianca Campagnari
Of course, research, discovery, and publication aren’t all scientists are required to do. Campagnari has also been initiated into the rigors of communicating her science and seeking funding for her work. She’s explained her research to an audience of friends, family, peers, and Museum staff during the Quasars to Sea Stars senior project presentations. She submitted information about her independent research to the Regeneron Science Talent Search—a nationwide contest among high school seniors run by the Society for Science & the Public since 1942—and was honored with a personal award of $2,000 and an award of $2,000 for her school (San Marcos High) to put towards STEM activities. In August, she’ll present her findings on methods for estimating species richness in micromollusks to other scientists and students at the World Congress of Malacology in Monterey. Most recently, she spoke about the value of the Quasars to Sea Stars program in front of Museum donors during our annual fundraising dinner, the Mission Creek Soirée.

Left to right: Director of Education Justin Canty, Quasars Diego Perez and Campagnari advocating for teen programs during the Mission Creek Soirée. Photos by Baron Spafford
Although the Quasars to Sea Stars program is naturally free for participating teens (and they earn money by working at the Museum), it costs money to run the program and support each quasar. Campagnari’s participation and research has been generously funded by ocean scientist and educator Laura Francis through the Sea Forward Fund. Other supporters of the Quasars to Sea Stars program include Edison International, Kirby-Jones Foundation, and the Donald and Coeta Barker Foundation. In addition to financial support, Campagnari received help and guidance not just from Geiger, but from Teen Programs Manager Jenna Rolle, Librarian Terri Sheridan (who has long played an active role in mentoring quasars), and her parents. Prior to her work generating the analysis on the shells, several UCSB students had contributed time to sorting samples as well.
Working collaboratively in a supportive environment and getting practice communicating about her work has given Campagnari the confidence to define her limits, as a good scientist should. When I heard her present her work during the senior project presentations last year, she said something that impressed me even more than her precocious publication record. Her presentation ended and the audience was invited to ask questions. One of her peers asked a good question about the behavior of one of the statistical models she compared. Campagnari clicked her slides back to show the audience what he was pointing out, and boldly said “I don’t know.” She went on to discuss the implications of the behavior, but those three words had already spoken volumes about her ethical need as a scientist to distinguish speculation from well-supported conclusions.
This tendency to beware assumptions and avoid overstatements is an important part of what makes science work. Campagnari explains: “I think saying ‘I don’t know’ definitely opens the door for more inquiry, which is important for learning things.” As demonstrated by her contribution to collective knowledge, she’s learned a great deal during her time here, thanks to her open curiosity. Campagnari didn’t know anything about micromollusks when she started working here, “but I became really passionate about them because it’s just cool to think about how there’s so much life that we can’t even see. They’re much more understudied than macrospecies. Anything that hasn’t been studied is interesting.”
This fall, she’ll follow her wide-ranging curiosity to University of Chicago, where she plans to study cellular molecular biology. “I’m really interested in molecular biology because it really ties all organisms together. We share a lot of the same processes, just across all of life.” Yet within that field, she’s flexible, and doesn’t yet have her sights on joining a particular lab. (Heads up, U. Chicago professors!) “I don’t really know specifically what I’m going to be interested in. What I’ve learned through quasars—through both working here [in IZ] and also everything else we’ve done in the program—is that you’ll learn about something in the morning, have never heard of it, and by the end of the day you’re completely obsessed with it and want to know more. So I’m not too worried about what I end up studying, because I know I’ll find it interesting after I spend enough time with it.” As far as we’re concerned, these are words to live by, whether you want to be a publishing scholar or simply a happy human being.
* Campagnari and Geiger (2018) “How many micromollusks are there? A case study on species richness in Hawai’i, with the description of a new species of Murdochella (Gastropoda: Epitoniidae).” The Nautilus 132 (3-4), pp. 83-90.
† Research Associate Chris Thacker, Ph.D.
‡ Geiger et al. (2007) “Techniques for collecting, handling, preparing, storing and examining small molluscan specimens” Molluscan Research 27 (1), pp. 1-50.






0 Comments
Post a Comment